Mechanisms of gene regulation by transcription factors
We explore the transcription regulatory mechanisms that govern genome regulation at the interface of transcription factors, chromatin proteins and the transcription machinery, framing findings in developmental processes.
Gene regulation is central to cellular destiny and heavily relies on the activity of transcription factors. Little is known about how transcription regulatory complexes assemble, how they negotiate chromatin and what underlies their functional specificity and diversity. Our projects aim at exploring the transcription regulatory mechanisms to ultimately gain insights into genome regulation. We focus on Hox transcription factors, a family of homeodomain transcription factors with key functions in development, evolution and physio-pathological processes.
Work from the team has contributed to clarify the Hox specificity paradox, by identifying the Hox intrinsic protein sequences responsible for specificity, and by discovering novel modes of interactions with the PBC class specificity cofactors. We also identified physical and functional links with chromatin proteins, including PcG and Mediator complex proteins, as well as links with the transcription pausing factor M1BP, connecting Hox protein activity to chromatin modification and to the activity of the basal transcription machinery.
More recently, we uncovered a novel facet of Hox protein function, where Hox proteins act in a paralogue non-specific manner, a property that better suits the shared Hox biochemical properties. Our current research directions aim at investigating at the physiological level how Hox specific and non-specific functions relate to each other, by exploring Hox protein functions in two tissues, the larval fat body and adult muscle development. Work also aims at uncovering molecular principles of Hox generic functions, which have so far not been studied.
Publications
M1BP is an essential transcriptional activator of oxidative metabolism during Drosophila development
Hox Proteins in the Regulation of Muscle Development
HoxB genes regulate neuronal delamination in the trunk neural tube by controlling the expression of Lzts1
The Generic Facet of Hox Protein Function
Hox functional diversity: Novel insights from flexible motif folding and plastic protein interaction
Hox proteins mediate developmental and environmental control of autophagy
M1BP is an essential transcriptional activator of oxidative metabolism during Drosophila development
Hox Proteins in the Regulation of Muscle Development
HoxB genes regulate neuronal delamination in the trunk neural tube by controlling the expression of Lzts1
Human ZKSCAN3 and Drosophila M1BP are functionally homologous transcription factors in autophagy regulation
Cooperation of axial and sex specific information controls Drosophila female genitalia growth by regulating the Decapentaplegic pathway
PLZF limits enhancer activity during hematopoietic progenitor aging
The Generic Facet of Hox Protein Function
Post-translational modifications of HOX proteins, an underestimated issue
Fattening the perspective of Hox protein specificity through SLiMming
Regulation of the positive transcriptional effect of PLZF through a non-canonical EZH2 activity
The Hox proteins Ubx and AbdA collaborate with the transcription pausing factor M1BP to regulate gene transcription
Hox functional diversity: Novel insights from flexible motif folding and plastic protein interaction
Hox Service Warranty Extends to Adult Bone Repair.
Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition).
A survey of conservation of sea spider and Drosophila Hox protein activities.
Cellular and molecular insights into Hox protein action.
A flexible extension of the Drosophila ultrabithorax homeodomain defines a novel Hox/PBC interaction mode.
Autophagy : Moving Benchside Promises to Patient Bedsides.
Plasticity versus specificity in RTK signalling modalities for distinct biological outcomes in motor neurons
Drosophila Hox transcription factors access the RNA Polymerase II machinery through direct homeodomain binding to a conserved motif of Mediator subunit Med19
Hox genes : a fertile interplay of concepts and methods.
Hox proteins mediate developmental and environmental control of autophagy
TAFA4, a Chemokine-like Protein, Modulates Injury-Induced Mechanical and Chemical Pain Hypersensitivity in Mice.
Distinct genetic requirements for BX-C mediated specification of abdominal denticles.
Deregulation of the Protocadherin Gene FAT1 Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy.
The emerging role of acetylation in the regulation of autophagy.
Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function.
Distinct molecular strategies for Hox-mediated limb suppression in Drosophila: from cooperativity to dispensability/antagonism in TALE partnership.
The MYST-containing protein Chameau is required for proper sensory organ specification during Drosophila thorax morphogenesis.
Hox proteins display a common and ancestral ability to diversify their interaction mode with the PBC class cofactors.
Insights into Hox protein function from a large scale combinatorial analysis of protein domains.
Characterisation of genome-wide PLZF/RARA target genes.
Pool-specific regulation of motor neuron survival by neurotrophic support.
Stable, conditional, and muscle-fiber-specific expression of electroporated transgenes in chick limb muscle cells
Selection of distinct Hox-Extradenticle interaction modes fine-tunes Hox protein activity.
Visualization of protein interactions in living Drosophila embryos by the bimolecular fluorescence complementation assay.
Control of DNA replication: a new facet of Hox proteins?
Reiterative use of signalling pathways controls multiple cellular events during Drosophila posterior spiracle organogenesis.
Regulation of Hox activity: insights from protein motifs.
The timing of emergence of muscle progenitors is controlled by an FGF/ERK/SNAIL1 pathway
The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation
Classification of sequence signatures: a guide to Hox protein function.
Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres
Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control.
A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax.
A molecular clock operates during chick autopod proximal-distal outgrowth
Liprin-alpha has LAR-independent functions in R7 photoreceptor axon targeting.
Chameau HAT and DRpd3 HDAC function as antagonistic cofactors of JNK/AP-1-dependent transcription during Drosophila metamorphosis.
Getting a molecular grasp on Hox contextual activity.
Control of the segmentation process by graded MAPK/ERK activation in the chick embryo
Hox-controlled reorganisation of intrasegmental patterning cues underlies Drosophila posterior spiracle organogenesis.
Ectopic Myf5 or MyoD prevents the neuronal differentiation program in addition to inducing skeletal muscle differentiation, in the chick neural tube
Tgfbeta signaling acts on a Hox response element to confer specificity and diversity to Hox protein function.
The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions.
Hox genes and apoptosis, from architecture to sculpture.
Reptin and pontin antagonistically regulate heart growth in zebrafish embryos.
A green fluorescent protein reporter genetic screen that identifies modifiers of Hox gene function in the Drosophila embryo.
The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression.
Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips
Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting.
Reconstitution of a functional core polycomb repressive complex.
A Drosophila Polycomb group complex includes Zeste and dTAFII proteins
DWnt4 and wingless elicit similar cellular responses during imaginal development.
Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes.
Notch signalling acts in postmitotic avian myogenic cells to control MyoD activation
Delta 1-activated notch inhibits muscle differentiation without affecting Myf5 and Pax3 expression in chick limb myogenesis
HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties
Paraxis is expressed in myoblasts during their migration and proliferation in the chick limb bud
dlarp, a new candidate Hox target in Drosophila whose orthologue in mouse is expressed at sites of epithelium/mesenchymal interactions
The Ste20 kinase misshapen regulates both photoreceptor axon targeting and dorsal closure, acting downstream of distinct signals
Dynamic expression of d-CdGAPr, a novel Drosophila melanogaster gene encoding a GTPase activating protein.
Distinct hox protein sequences determine specificity in different tissues.
pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila
nessy, an evolutionary conserved gene controlled by Hox proteins during Drosophila embryogenesis
Antagonist activity of DWnt-4 and wingless in the Drosophila embryonic ventral ectoderm and in heterologous Xenopus assays.
SSX and the synovial-sarcoma-specific chimaeric protein SYT-SSX co-localize with the human Polycomb group complex.
Identification of the t(15;17) in AML FAB types other than M3: evaluation of the role of molecular screening for the PML/RARalpha rearrangement in newly diagnosed AML. The Medical Research Council (MRC) Adult Leukaemia Working Party
SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation
The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain
Wnt and TGFbeta signals subdivide the AbdA Hox domain during Drosophila mesoderm patterning.
RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor.
Drosophila Hox complex downstream targets and the function of homeotic genes.
Hox genes in evolution: protein surfaces and paralog groups.
The Drosophila teashirt homeotic protein is a DNA-binding protein and modulo, a HOM-C regulated modifier of variegation, is a likely candidate for being a direct target gene.
Inhibition of mitogen-induced DNA synthesis by bafilomycin A1 in Swiss 3T3 fibroblasts
wingless and DWnt4, 2 Drosophila Wnt genes, have related expression, regulation and function during the embryonic development.
Genetic and molecular analysis of terminal deletions of chromosome 3R of Drosophila melanogaster.
DWnt-4, a novel Drosophila Wnt gene acts downstream of homeotic complex genes in the visceral mesoderm.
The modifier of variegation modulo gene acts downstream of dorsoventral and HOM-C genes and is required for morphogenesis in Drosophila.
A quick method for immunoscreening recombinant bacterial colonies.
Cell lineage-specific expression of modulo, a dose-dependent modifier of variegation in Drosophila.
Homeotic control in Drosophila; the scabrous gene is an in vivo target of Ultrabithorax proteins.
News
We’re pleased to share some great news about our researchers’ achievements! Several projects from our teams have been selected for funding by the ANR and FRM, highlighting their hard work and innovative research.
Recent achievements and promotions
Join the IBDM for your internship!
Seeking for your Master internship? The IBDM seems like the right place to do it? Check out our offers.
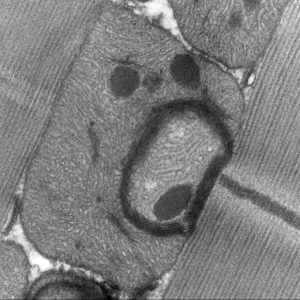
The Saurin/Graba group discover the transcription factor M1BP as new major regulator of oxidative metabolism.
The team of Yacine Graba and Andrew Saurin identified an atypical function for HoxB genes during spinal cord development.
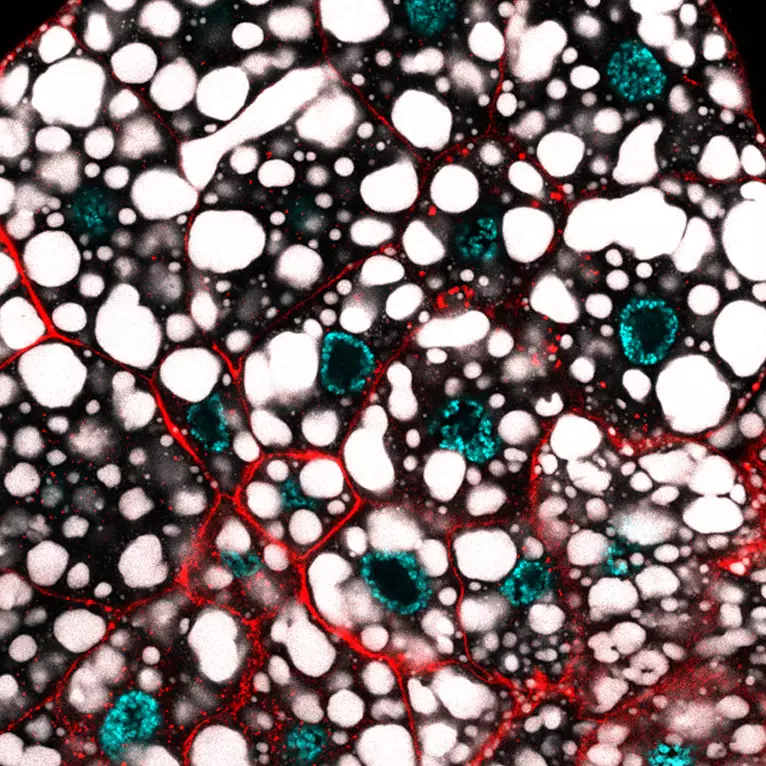
How two recently discovered and oppositely acting transcriptional regulators control metabolism in the Drosophila larval fat body, with a special attention on metabolic paths linked to fat accumulation.
The work will investigate how transcription factor protein sequences influence LLPS, nuclear sub-compartmentalization and gene regulation during development.
The project aims at uncovering the molecular and cellular bases for Ubx and AbdA distinct motif usage.
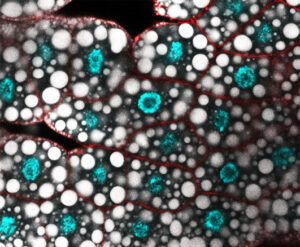
The M2 project will use state-of-the-art genomics profiling techniques, Drosophila genetics, imaging and molecular biology for studying the antagonistic transcriptional control and dysfunction of fat body lipid metabolism development.
The M2 project will use state-of-the-art genomics profiling techniques, Drosophila genetics, imaging and molecular biology for studying muscle development.
Team members
Alumni
Funding bodies