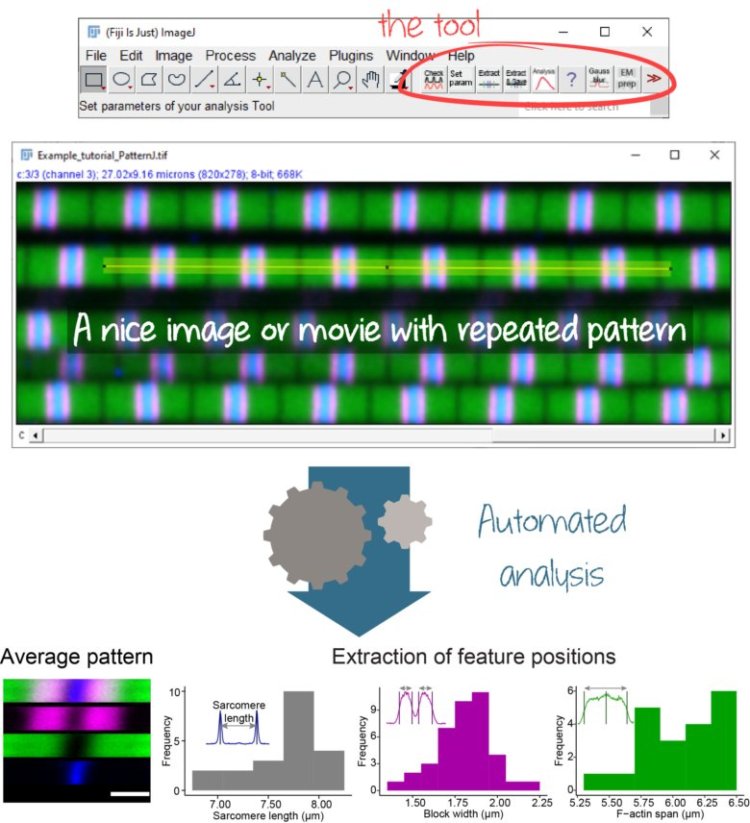
Regular spatial patterns are ubiquitous forms of organization in nature. In animals, regular patterns can be found from the cellular scale to the tissue scale, and from early stages of development to adulthood. To understand the formation of these patterns, how they assemble and mature, and how they are affected by perturbations, a precise quantitative description of the patterns is essential. However, accessible tools that offer in-depth analysis without the need for computational skills are lacking for biologists.
In a new article published in Biology Open, Mélina Baheux Blin, Vincent Loreau, Frank Schnorrer, and Pierre Mangeol present PatternJ, a novel toolset to analyze regular patterns precisely and automatically. This toolset, to be used with the popular imaging processing program ImageJ/Fiji, facilitates the extraction of key geometric features within and between pattern repeats in static images and time-lapse series. PatternJ was tested with simulated data and the authors showed it is useful on images of sarcomeres from insect muscles and contracting cardiomyocytes, actin rings in neurons, and somites from zebrafish embryos obtained using confocal fluorescence microscopy, STORM, electron microscopy, and brightfield imaging.
PatternJ’s straightforward use and functionalities make it valuable for various scientific fields requiring quantitative one-dimensional pattern analysis, including the sarcomere biology of muscles or the patterning of mammalian axons, speeding up discoveries with the bonus of high reproducibility.