Polarization and binary cell fate decisions in the nervous system
We are analyzing how the divisions of neuronal progenitors are regulated and how differentiated neurons are produced in a robust manner.
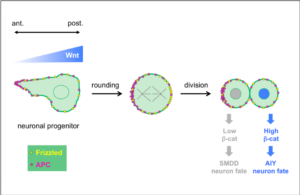
Neurons are often generated by asymmetric divisions of neuronal progenitors such as neural stem cells. During this process, a progenitor cell divides asymmetrically to generate two neurons with different identities, or one neuron and a new progenitor. In the nervous system, the asymmetric divisions of the different progenitors are tightly coordinated, implying a communication between cells. In addition, during nervous system development, a very precise set of different neuronal types is produced, meaning that their specification process has to be very robust.
Our team analyzes how the asymmetric divisions of neuronal progenitors are controlled and how various differentiated neuron types are produced in a robust manner. To address these questions we are using the nematode C. elegans as a model organism. C. elegans is a good system to study this process as its nervous system is simple and well characterized. In addition, in C. elegans, the lineage history of every neuron is known, the embryos are transparent and their development can be easily followed by 4D-videomicroscopy. The C. elegans system also offers numerous tools to dissect the molecular basis of biological processes such as genome-wide screens, transgenesis or CRISPR genome engineering. The combination of genome engineering and quantitative live imaging allows us to follow the dynamics of proteins and gene expression in vivo during nervous system development.
By characterizing the mechanisms controlling neuronal progenitor divisions and differentiation, our work may have an impact on the development of treatments against some types of cancer or neurodegenerative diseases.
Publications
Left/right asymmetrically expressed ephrin and Flamingo proteins regulate lateralized axon growth in C. elegans.
PRC1 chromatin factors strengthen the consistency of neuronal cell fate specification and maintenance in C. elegans
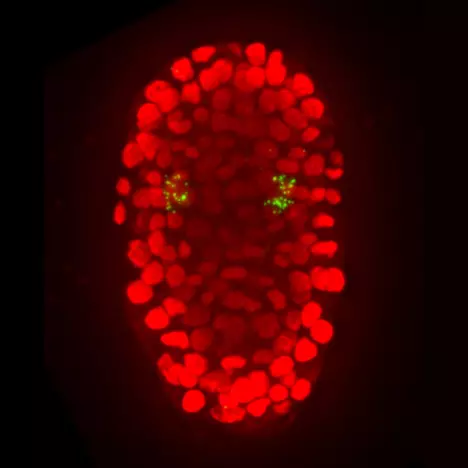
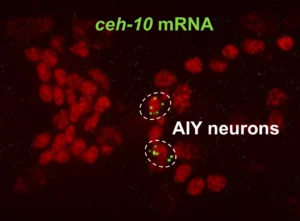
Multiple neural bHLHs ensure the precision of a neuronal specification event in Caenorhabditis elegans
Imaging of native transcription and transcriptional dynamics in vivo using a tagged Argonaute protein
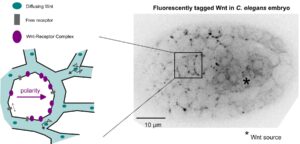
Wnt ligands regulate the asymmetric divisions of neuronal progenitors in C. elegans embryos
Atypical transcriptional activation by TCF via a Zic transcription factor in C. elegans neuronal precursors_edited
Left/right asymmetrically expressed ephrin and Flamingo proteins regulate lateralized axon growth in C. elegans.
Transfer of polarity information via diffusion of Wnt ligands in C. elegans embryos
Labelling of Active Transcription Sites with Argonaute NRDE-3-Image Active Transcription Sites in vivo in Caenorhabditis elegans
PRC1 chromatin factors strengthen the consistency of neuronal cell fate specification and maintenance in C. elegans
Multiple neural bHLHs ensure the precision of a neuronal specification event in Caenorhabditis elegans
Imaging of native transcription and transcriptional dynamics in vivo using a tagged Argonaute protein
Neuronal specification in C. elegans: combining lineage inheritance with intercellular signaling
Wnt ligands regulate the asymmetric divisions of neuronal progenitors in C. elegans embryos
Zic Genes in Nematodes: A Role in Nervous System Development and Wnt Signaling
Zic-proteins are repressors of dopaminergic forebrain fate in mice and C. elegans
β-catenin-driven binary cell fate decisions in animal development
How targets select activation or repression in response to Wnt_edited
Atypical transcriptional activation by TCF via a Zic transcription factor in C. elegans neuronal precursors_edited
Setting-up a simple light sheet microscope for in toto imaging of C. elegans development
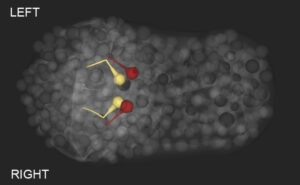
Notch-dependent induction of left/right asymmetry in C. elegans interneurons and motoneurons.
Lineage programming: navigating through transient regulatory states via binary decisions.
Analysis of multiple ethyl methanesulfonate-mutagenized Caenorhabditis elegans strains by whole-genome sequencing
Wnt asymmetry and the terminal division of neuronal progenitors.
Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans.
News
Hidden asymmetries in the brain
And yet it diffuses!
Join the IBDM for your internship!
Seeking for your Master internship? The IBDM seems like the right place to do it? Check out our offers.
The Bertrand team shows that PRC1 chromatin factors play a role in the robustness of neuronal fate against gene expression noise and environmental perturbations.
7 IBDM teams have received grants from ANR
7 IBDM teams have received grants from the Agence Nationale pour la Recherche (ANR) in 2021. Congratulations to Vincent Bertrand, Harold Cremer, Pascale Durbec, André Le Bivic, Pierre-François
Antoine Barrière and Amel Toudji-Zouaz in the Bertrand team developed a new method to image and quantify transcription in vivo.
In a collaborative study published in Development, the teams of Vincent Bertrand and Pierre-François Lenne analyze the role played by Wnt ligands in the divisions that generate neurons during nervous system development.
During this master project (M1 or M2), the student will analyze the dynamics of PRC1 dots during neuronal specification and maintenance by in vivo time-lapse imaging, and characterize the nature of these dots.
PhD position on the robustness of cell fate specification programs – Bertrand Lab (June 2022)
A PhD position on the robustness of cell fate specification programs – Bertrand Lab (June 2022) at the Developmental Biology Institute of Marseille (IBDM), France, in the group